March 15, 2018
CRISPR then, now and in the future — three current and past NU scientists with multiple links to CBC: Elizabeth McNally, Navdeep Chandel and Eric Sontheimer
In the article cited below, originally published in the Northwestern Medicine Magazine, Will Doss describes the parallels between the local (Northwestern-centric) and global history of CRISPR Cas9 gene editing methodology. Doss takes a reader on a captivating journey, full of fascinating examples and anecdotes, from the time of CRISPR discovery to futuristic applications that carry a potential to eradicate diseases that are currently untreatable. Among the NU scientists who were taken by the “CRISPR storm” in their research and are featured in the article are three CBC scientists: Elizabeth McNally, Navdeep Chandel and Eric Sontheimer (at UMMS now). Enjoy the read and, following the article, check out the impressive list of the ties these three scientists have with CBC! Congratulations to all featured researchers!
The CRISPR revolution
Northwestern Medicine scientists usher in a new era of genetic research.
Northwestern Medicine Magazine | by WILL DOSS | March 9, 2018
The McNally lab members were shocked.
For the first time, they had used a new gene editing technique that promised to transform how scientists investigated the human genome. Prior to that day in late 2014, editing genes in mammalian cells had been a time-consuming process — sometimes requiring screening hundreds of clones to find one with altered DNA — and it often ended in failure. But then, Eugene Wyatt, PhD, a postdoctoral fellow in the lab, generated a cellular model of genetic disease in human embryonic kidney cells faster than ever before by harnessing a specialized region of DNA called CRISPR.
“More than 80 percent of the clones showed evidence of editing,” says Elizabeth McNally, MD, PhD, the Elizabeth J. Ward Professor of Genetic Medicine and director of Feinberg’s Center for Genetic Medicine. “This was truly revolutionary — older methods only worked in very specific cells and relied on waiting for a cell’s natural machinery to edit genes. With CRISPR, that machinery could be directly introduced into the cells, dramatically improving efficiency.”
Tasks such as creating a mouse model of disease, which previously took about three years, now would take just four to six months, allowing scientists around the world to more quickly understand mechanisms of disease and more efficiently translate those discoveries from bench to bedside. At Feinberg, CRISPR gene editing is being used today in many settings, including to isolate mutations that cause neurological diseases and to run large-scale genetic screens to understand how individual genes can damage — or protect — cells.
Northwestern Medicine scientists are quick to point out that CRISPR-Cas9 is not just a laboratory tool; it also has tremendous potential for use in patients, though there are still unresolved ethical and regulatory issues to think through before editing live human genomes. But its first experimental use in patients is closer than most realize, according to McNally.
“If you had asked me last year, I would’ve told you it will be ten years before somebody injects a patient with a genome-editing virus,” McNally says. “Now I think it’s about two or three years away.”
That prediction underlines the head-spinning pace at which CRISPR technology is changing genetics. The first peer-reviewed papers showing successful gene editing in mammals were published just five years ago.
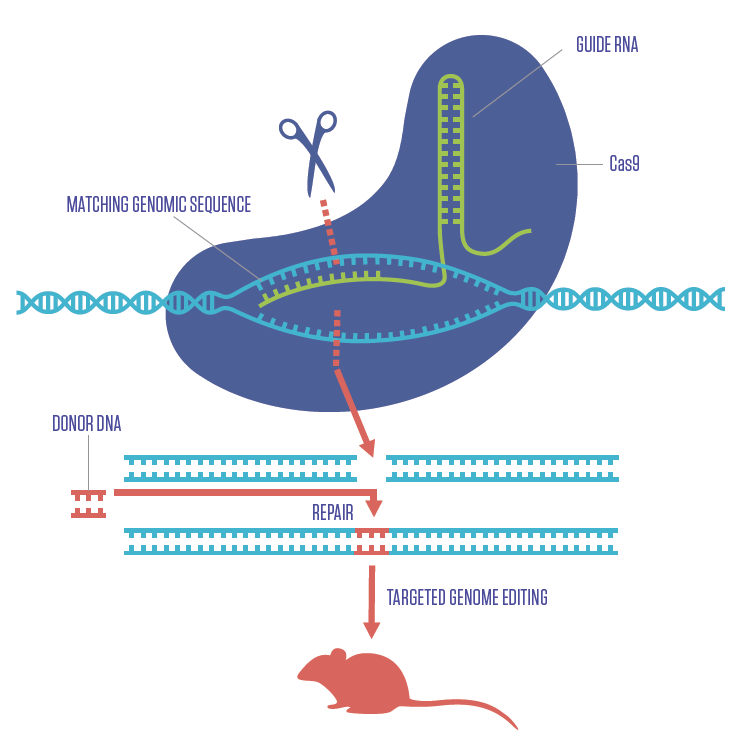
HOW CRISPR WORKS
Just as CRISPR was transforming what scientists can do with cells, it equally changed the landscape for genetically engineering mice to model human diseases. The diagram here illustrates how the CRISPR-Cas9 gene editing complex works. A sequence of RNA (green) programmed to find a specific segment of DNA (blue) is inserted into a cell along with the Cas9 enzyme (purple). Once the RNA finds the matching DNA, Cas9 cuts it. In that space, a fragment of donor DNA containing new genetic information (red) can be inserted.
The Center for Genetic Medicine houses the Transgenic and Targeted Mutagenesis Laboratory (TTML) directed by Lynn Doglio, PhD. Under the guidance of Raj Awatramani, PhD, the TTML Core Facility has embraced using CRISPR gene editing to make novel and critically essential models to accelerate research. Pei-Ken Hsu, PhD, joined the TTML core in 2016 to further expand gene editing in the TTML Core, where he has now worked with many investigators across Northwestern.
CRISPR AT FEINBERG
Using CRISPR to edit the genomes of human cells or model organisms has become a staple of research activities at Feinberg — especially when combined with induced pluripotent stem cells (IPSCs).
Scientists can harvest adult cells from a human subject, turn those into stem cells and direct the resulting IPSCs to develop into a specific type of tissue, such as cardiac cells, neurons or skeletal muscle tissue. This technique helps investigators identify cellular mechanisms of disease and can be combined with CRISPR to isolate disease-causing mutations.
For example, stem cells can be created using genetic material from patients with amyotrophic lateral sclerosis (ALS), and those cells differentiated into motor neurons. Investigators can compare those neurons to neurons from healthy individuals, looking for genetic mutations in the diseased model. If they find a suspicious mutation, they can use CRISPR to reverse the genetic mutation, creating a stem cell line identical to the patients’ cells, but without the mutation — this is called an isogenic control line.
“This allows us to test whether a phenotype or defect in a motor neuron is caused by that particular mutation,” says Evangelos Kiskinis, PhD, assistant professor of Neurology in the Division of Neuromuscular Disease. “If the defect goes away, we’ve established the mutation is necessary for the defect.”
Kiskinis also uses CRISPR to do the opposite experiment. He takes a healthy stem cell line, introduces the mutation using CRISPR and asks if that change is sufficient enough to induce the same phenotype. This method becomes even more important when looking at diseases caused by a combination of genes, such as epilepsy. Kiskinis and his colleagues have introduced a known epilepsy-causing gene variant into a variety of stem cell lines derived from healthy individuals. They’re looking for insights into the impact of an individual’s broader genetic background and why certain people develop epilepsy and others don’t.
“This is the first time we are able to do this in human cells,” Kiskinis says. “Before, it was technically possible, but extremely challenging; it would take a very long time.”
Another application of CRISPR is in large-scale genetic screens, a fast and simple way to investigate the effects of individual genes in cells. Navdeep Chandel, PhD, the David W. Cugell, MD, Professor of Medicine in the Division of Pulmonary and Critical Care Medicine, used this method to narrow down which genes made a person vulnerable to Parkinson’s disease after repeated exposure to an herbicide called paraquat. Epidemiological studies indicate that farmers exposed to paraquat have a higher risk of Parkinson’s disease. A major cause of Parkinson’s disease is a loss of function in dopamine neurons, which are known to be vulnerable to oxidative stress.
“Paraquat generates a lot of oxidants. Naturally those dopamine neurons will be the most susceptible to damage from the pesticide,” says Chandel. His team conducted a CRISPR positive-selection screen, creating thousands of cells with one individual gene turned off. They then exposed the cells to paraquat — the majority of them died, but not all. Certain cells with knocked-out genes were resistant to paraquat, suggesting those genes may be responsible for the toxicity. One gene in particular, called POR, was pinpointed as the main source of damage-causing oxidation, according to Chandel.
Investigating oxidant stress could pay dividends in the future, including in the development of drugs designed to generate oxidative stress in cancer cells, killing them while leaving healthy cells alone. While some drugs currently exist, not enough is known about their pathways to create a functioning compound, says Chandel, a member of the Robert H. Lurie Comprehensive Cancer Center of Northwestern University.
“The biology of oxidative stress is still a mystery,” he says. “CRISPR positive-selection screens could be a way to figure it out.”
UNANSWERED QUESTIONS
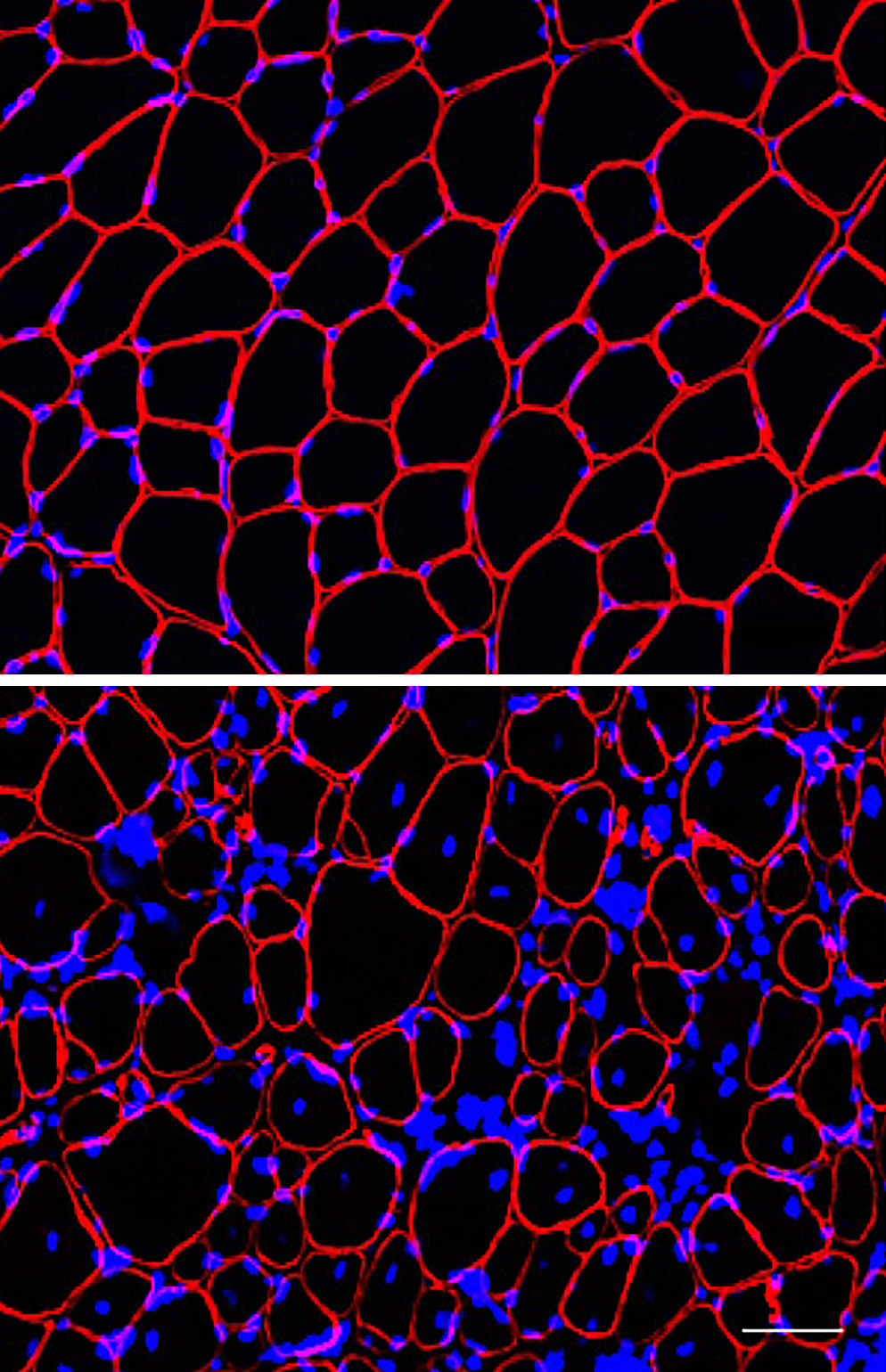
Human induced pluripotent stem cells that have been gene edited with CRISPR from the lab of Elizabeth McNally, MD, PhD.
While there’s little legal or ethical debate around using CRISPR with cultured cells or non-embryonic stem cells, using the tool to edit the genome of living humans — especially in a manner where changes would be inherited by children — is still uncharted territory. Scientists in China reported using CRISPR to edit a gene responsible for a deadly blood disorder in non-viable embryos, but few embryos survived and it set the international scientific community off into a fiery debate.
However, it’s no longer a question of if, but rather a question of when CRISPR-like technology will be used in humans, according to Raj Awatramani, PhD, associate professor of Neurology in the Division of Movement Disorders.
“Society doesn’t keep pace with the science,” says Awatramani, who uses CRISPR to create model neurons vulnerable to Parkinson’s disease. “We need to have new ethical guidelines to deal with genome editing in humans. Right now, it’s gray, gray and grayer.”
Several companies are racing to be the first to use CRISPR in living humans, usually to treat serious chronic diseases such as HIV/AIDS or Duchenne muscular dystrophy, according to McNally. For Duchenne, the proposed treatments would involve injecting a patient — probably a young child — with a virus containing CRISPR material. Early models show that even if uptake only occurs in some cells, symptoms are relieved because the corrected cells tend to compensate for the diseased cells.
“It’s nerve-wracking when we’re talking about children,” says McNally, who testified about the importance of using this method to treat genetic disease to the U.S. House of Representatives Committee on Science, Space and Technology in 2015. “Diseases like Duchenne muscular dystrophy are so difficult for patients and their families that it could be argued trying the therapy is the right thing to do, as long as it’s reasonably safe.”
Regardless of its future as a therapeutic tool, CRISPR has entrenched itself as a central mechanism for genetics research, in an astonishingly short period of time. Combined with the adaptability of IPSC, the technologies have irrevocably changed genetics for the better.
“There’s no question, when people look back 100 years from now, they’ll find it hard to believe all of this was discovered at the same time,” McNally says. “This is truly a revolutionary time for genetics.”
LOCAL BEGINNINGS
First discovered byFrancisco Mojica in Spain in 1993, CRISPR is made up of short, repeated sequences of DNA and non-coding spacer DNA. Its purpose confounded investigators. But by 2003, Mojica had identified thousands of sequences of genetic code in CRISPR that matched snippets of bacterial and viral genomes. He hypothesized that CRISPR was part of an adaptive immune system that copies sequences from invading microbes to ward off viral infections of bacteria. Scientists around the world recognized CRISPR’s potential as an experimental tool and set out to investigate it further.
Northwestern University was the setting of important research establishing the basic science behind CRISPR. In 2008, Luciano Marraffini, PhD, a postdoctoral fellow in the lab of Erik Sontheimer, PhD, then an associate professor of Biochemistry, Molecular Biology and Cell Biology at Northwestern’s Weinberg College of Arts and Sciences, became the first to empirically prove CRISPR destroys plasmid or virus DNA molecules directly and to suggest that it can be programmed to target any DNA. They published their findings in a landmark paper in Science.
“Our work was a breakthrough in the understanding of CRISPR, since it explained how it works at the molecular level,” says Marraffini.
Sontheimer adds, “Most important of all, we were the first to recognize and explicitly articulate the possibility that CRISPR could be repurposed for genomic engineering.”
The pair filed a patent declaring CRISPR could be used to manipulate the genomes of complex organisms, but the patent was denied, citing lack of experimental demonstration.
By then, other investigators had linked the CRISPR system with Cas9, an enzyme that modifies DNA, and begun to harness the whole complex for genome editing. The final breakthrough happened in January 2013, when five groups from around the world published independent studies within three weeks showing the system could be programmed to target specific points of DNA in mammalian cells.
Among them were scientists from the Massachusetts Institute of Technology, who collaborated with Marraffini, then at The Rockefeller University, to publish a paper in Science demonstrating that the CRISPR sequence can be transcribed into short RNA sequences that drag Cas9 to a specific locus and cut the DNA, turning the targeted gene off.
This discovery rocked the field of genetics: Just nine months after the initial Science publication, an additional 1,500 articles on the CRISPR-Cas9 complex had been published, refining and improving the tool. This was made possible by the decision to make CRISPR reagents readily available online with instructions to help scientists design the right experimental tools, says McNally.
Source:
Adapted (with modifications) from Northwestern Medicine Magazine, by Will Doss, posted on March 9, 2018.
McNalle, Chandel and Sontheimer’s ties to CBC:
▸ Catalyst Review Board (CRB)
Navdeep Chandel (NU) – CRB Board Member (2014-present)
CBC HTS Award (2015):
PIs: Navdeep Chandel and Inma Martinez-Reyes (NU)
▸ Discovering Small Molecules to Improve Mitochondrial Function for Neuron Protection
CBC Scholars “Loop Connections” Seminar (2012):
▸ Processing-Independent CRISPR RNAs Limit Natural Transformation in Neisseria meningitidis
Erik Sontheimer (UMMS, at NU then) – Invited Seminar Speaker
▸ CBC Science Day (2011):
Erik Sontheimer (UMMS, at NU then) – Science Day Speaker
CBC Catalyst Award (2011):
PIs: David Eddington and Jalees Rehman (UIC), and Navdeep Chandel (NU) for the project:
▸ Oxygen Sensing in Endothelial Progenitor Cells
CBC Spark Award (2009):
Erik Sontheimer (UMMS, at NU then), Alexander Mankin (UIC) and Jonathan Staley (UChicago) for the project:
▸ Noncoding RNA Structure, Function, and Evolution
CBC Spark Award (2008):
PIs: Robert Goldman and Jonathan Widom (NU), Stephen Kron and Harindar Sighn (UChicago) and Elizabeth McNally (NU now, UChicago then) for the project:
▸ The Chicago Laminome Project
CBC Catalyst Award (2007):
PIs: Elizabeth McNally (NU now, UChicago then), P. Schumacker and H. Ardehali (NU), and R. J. Solaro (UIC) for the project:
▸ Proteomic Analysis of Mitochondrial and Sarcomeric Proteins in Cardiomyopathy
CBC Catalyst Award (2007):
PIs: Erik J. Sontheimer (UMMS, at NU then), Jonathan P. Staley and Manyuan Long (UChicago) for the project:
▸ Gene Regulation by Natural Antisense RNAs in Yeast
Spark Review Panel:
Erik J. Sontheimer (UMMS, at NU then) – Spark Reviewer